This manual gives you a walk-through on how to use the logP Plugin:
Introduction
The logP Plugin calculates the octanol/water partition coefficient, which is used in QSAR analysis and rational drug design as a measure of molecular hydrophobicity. The calculation method is based on the publication of Viswanadhan et al. The logP of a molecule is composed of the increment of its atoms. However, the algorithm described in the paper was modified at several points:
- Many atomic types were redefined to accommodate electron delocalization and contributions of ionic forms were added.
- The logP of zwitterions are calculated from their logD value at their isoelectric point.
- The effect of hydrogen bonds on logP is considered if there is a chance to form a six membered ring between suitable donor and acceptor atoms.
- New atom types were introduced especially for sulfur, carbon, nitrogen and metal atoms.
To find details on logP calculation, see the following page.
The result of the calculation appears in a new window, either in a MarvinView (for a 2D view) window or in a MarvinSpace (for a 3D view) window.

Fig. 1 LogP result window with atomic increments displayed in MarvinView
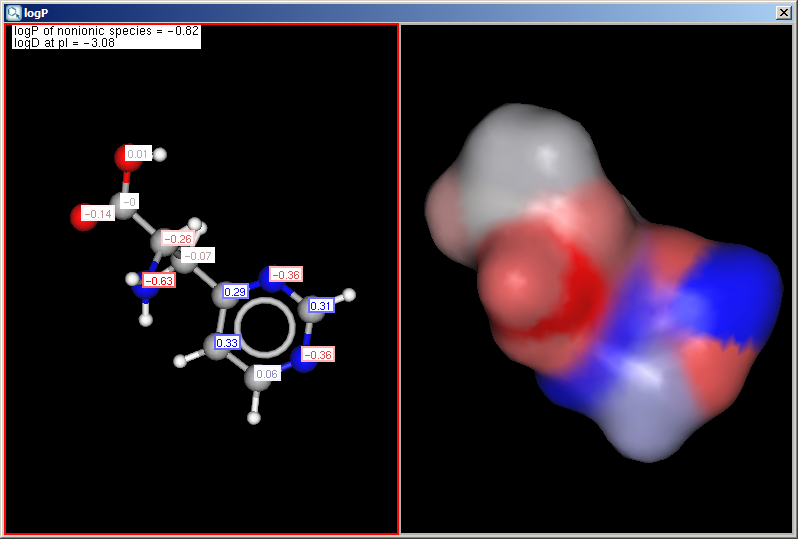
Fig. 2 LogP result window with atomic increments displayed in MarvinSpace
Options
General Options
Different general options can be set in the logP Options window:
- Method
- VG: the calculation method derived from Viswanadhan et al. is applied.
- KLOP: logP data from Klopman et al. is applied.
- PHYSPROP: logP data from PHYSPROP© database is used.
- User defined: if a training set of structures and corresponding experimental logP values is available, it can be used as a database for logP calculations.
See the manual page on creating such training sets. - Weighted: default setting. The use of methods can be combined by the user. Selecting this method turns the Method weights section active.
- Training ID: if the User defined or the Weighted method is selected, this dropdown list becomes active. All created training sets are listed here. Choose the one to apply for the calculation.
- Method weights: you can set the weights of the methods used in the calculations. Acitve only if Weighted method is selected.
- Electrolyte concentration
- Cl- concentration: can be set between 0.1 and 0.25 mol/L.
- Na+ K+ concentration: can be set between 0.1 and 0.25 mol/L.
- Take major tautomeric form: the logP of the major tautomer will be calcutated.
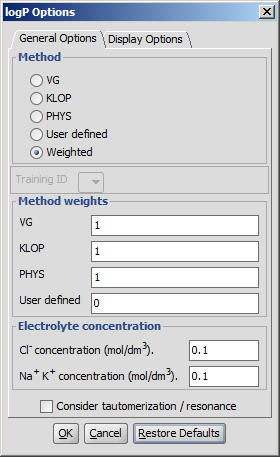
Fig. 3 logP Options window with the General Options panel
Display Options
Different display options can be set in the logP Options window:
- Decimal places: setting the number of decimal places with which the result value is given.
- Show value
- Increments: calculates and shows the increments for atoms one by one.
- logP : shows the value of logP
- Display in MarvinSpace: the result window opens as 3D MarvinSpace viewer. If unchecked, the results will be shown in a 2D MarvinView panel.
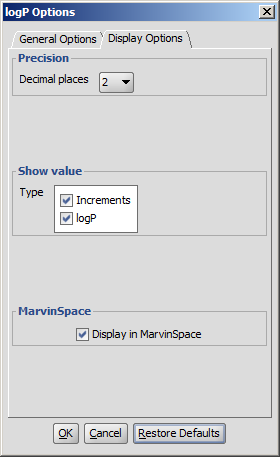
Fig. 4 logP Options window with the Display Options panel
References
- Viswanadhan, V. N.; Ghose, A. K.; Revankar, G. R.; Robins, R. K., J. Chem. Inf. Comput. Sci., 1989, 29, 163-172; doi
- Klopman, G.; Li, Ju-Yun.; Wang, S.; Dimayuga, M.: J.Chem.Inf.Comput.Sci., 1994, 34, 752; doi
- PHYSPROP© database
- Csizmadia, F; Tsantili-Kakoulidou, A.; Pander, I.; Darvas, F., J. Pharm. Sci., 1997, 86, 865-871; doi