Introduction
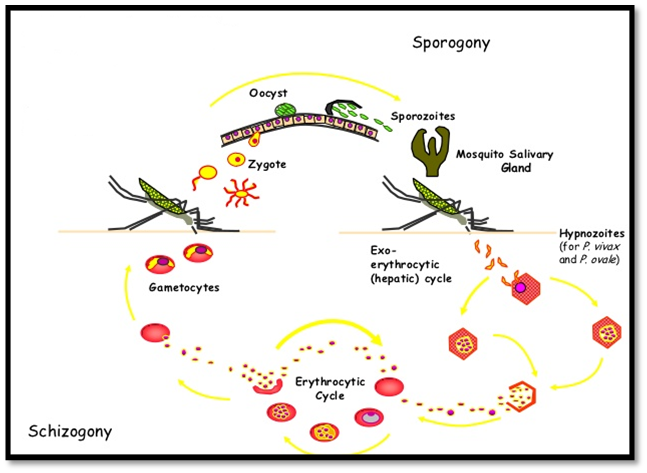
Plasmodium vivax is one of the five species of malaria parasites that commonly infect humans and the most common cause of frequent malaria. It is less virulent than Plasmodium falciparum, the deadliest of the five, but vivax malaria can lead to severe disease and death. P. vivax is carried by the female Anopheles mosquito. The genome is 22 MB in size; the GC content is 45% (unlike some other Plasmodia sequences, which have notably low GC content) and about 5000 genes have been predicted. (Carlton et al., 2001). P. vivax nuclear DNA is distributed between 14 linear chromosomes that range in size from 1.2 to 3.5 Mb (Carlton, et al., 1999). Currently we have overall only 1% experimental protein structures in PDB.