|
Chemgenome software package | To know about Genome Analysis | To know more about Genome Analysis | Go to Genomics Tutorial
Genome analysis entails the prediction of genes in uncharacterized genomic sequences. The objective is to be able to take a newly sequenced uncharacterized genome and break it up into introns, exons, repetitive DNA sequences, transposons etc. and other elements.
The various components of Genome Analysis are:
Gene Evaluation: Given a DNA sequence, what part of it codes for a protein and what part of it is junk DNA.
Genome Classification: Classify the junk DNA as intron, untranslated region, transposons, dead genes, regulatory elements etc.
Gene Prediction: Predict the coding regions in a newly sequenced genome into the genes (coding) and the non-coding regions.
Importance of Genome Analysis ?
Several genetic disorders like Huntington’s disease, Parkinson’s disease, sickle cell anemia etc. are caused due to mutations in the genes or a set of genes inherited from one generation to another. There is a need to understand the cause for such disorders. An understanding of the genome organization can lead to concomitant progresses in drug-target identification.
Comparative genomics has become a very important emerging branch with tremendous scope, for the above mentioned reasons. If the genome for humans and a pathogen, a virus causing harm is identified, comparative genomics can predict possible drug-targets for the invader without causing side effects to humans.
Over the past two decades genetic modification has enabled plant breeders to develop new varieties of crops like cereals, soya, maize at a faster rate. Some of these called as transgenic varieties have been engineered to possess special characteristics that make them better. Recently efforts are on in the area of utilizing GM (genetically modified) crops to produce therapeutic plants.
Genome Analysis is also important in SNP (Single nucleotide polymorphisms) discovery and analysis. SNPs are common DNA sequence variations that occur when a single nucleotide in the genome sequence is changed. SNP occurs every 100 to 300 bases along the human genome. The SNP variants promise to significantly advance our ability to understand and treat human diseases.
Mice and humans contain roughly the same number of genes – about 28,000 protein coding regions. The chimp and human genomes vary by an average of just 2% i.e. just about 160 enzymes. What is in the nature of these genes that confer such a huge phenotypical difference to these organisms?
The genome projects will have additional benefits that at present can only be guessed at. For e.g. we think that most of the intergenic DNA has no function, but perhaps this is because we do not know enough about it. There is one final reason for genome projects. The work stretches current technology to its limits. Genome analysis therefore represents the frontier of molecular biology, territory that was inaccessible just a few years ago.
What are the various comparison parameters??
True positives (TP): Genes evaluated as genes.
False positives (FP): Non-genes evaluated as genes.
True negatives (TN): Non-genes evaluated as non-genes.
False negatives (FN): Genes evaluated as non-genes.
Actual Positives(AP) = TP+FN.
Actual negatives (AN) = FP+TN.
Predicted number of positives (PP) =TP+FP.
Predicted number of negatives (PN) = TN+FN.
Sensitivity (SS) =TP / (TP+FN).
Specificity (SP) =TP / (TP+FP).
Correlation-Coefficient (CC) = 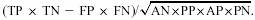
Qualitatively speaking, Sensitivity is the ability to identify as many correct genes as possible. Specificity is measure of the proportion of correct genes out of the total genes identified.
What is Chemgenome? How is it different from other softwares?
Chemgenome is an ab-initio gene evaluation and prediction software that uses physico-chemical properties to construct a 3D vector to answer the fundamental question ,”What is a gene” ?