 |
|
| Protein Structure Prediction |
Considered to be the holy grail of modern biology, a solution to the protein folding problem has enormous potentially beneficial impact on society. Prediction of three dimensional structures of drug targets, design of biocatalysts and nanobiomachines are a few of the multitude of foreseeable applications. At IIT Delhi, we have been developing a computational protocol for modeling and predicting protein structures at the atomic level. The software is named Bhageerath after the great Indian king who managed to accomplish the impossible task of getting the Ganges from heaven to earth. |
 |
 |
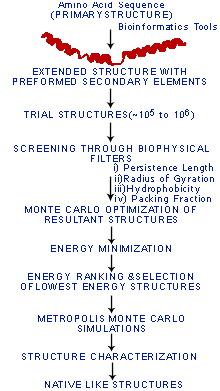
A computational pathway for bracketing native-like structures of small alpha helical globular proteins.
|
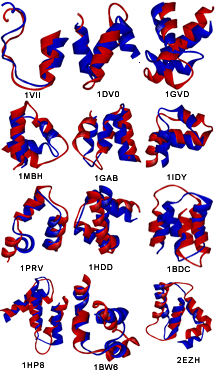
The lowest RMSD structure emerging from the proposed pathway superimposed on the corresponding native structure for each of the twelve test proteins: Native is represented in blue and the predicted native-like structure in red.
|
REFERENCE
1. Jayaram, B., Bhushan, K., Shenoy, S. R., Narang, P., Bose, S., Agrawal, P., Sahu, D., Pandey, V.S. Bhageerath : An Energy Based Web Enabled Computer Software Suite for Limiting the Search Space of Tertiary Structures of Small Globular Proteins. Nucl Acids Res., 2006. , 34, 6195-6204; doi: 10.1093/nar/gkl789 [Full Paper]
2. Narang, P., Bhushan, K., Bose, S., and Jayaram, B. Protein structure evaluation using an all-atom energy based empirical scoring function, J. Biomol.Str.Dyn, 2006 23, 385-406. [ABSTRACT]
3. Narang, P., Bhushan, K., Bose, S. and Jayaram, B. A computational pathway for
bracketing native-like structures for small alpha helical globular proteins. Phys. Chem. Chem. Phys., 2005 7, 2364-2375. 
| S.NO. |
Software |
Description |
Version |
1 |
|
Predicts native-like structures for small globular proteins |
alpha |
2 |
|
Calculates intramolecular energy of a protein in component-wise break up. |
1.0 |
3 |
|
Energy minimizer for proteins |
1.0 |
|
|
|
|

