Molecule Graph
Graphs offer a natural way of representing chemical structures.
In this case the atoms are the nodes of the graph and the bonds
are the edges. We can then “color” the nodes with atom types and
“weigh” the edges with bond types.
This graph, which contains nodes and edges with different properties,
is the molecular graph.
Elements of the molecule graph are:
Only one connection/bond is allowed between any two atoms and no self
connection is allowed. To represent this molecule graph in a computer
the following matrices are introduced:
- Connection table, ctab:
the i-th row defines the indices of the atoms connected to the
i-th atom.
|
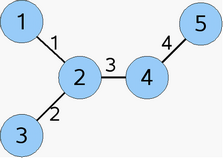 |
Connection table (ctab):
[2]
[1, 3, 4]
[2]
[2, 5]
[4]
|
- Bond table, btab:
bond indexes between atom i and atom j.
For small molecules it is a square matrix, for large molecules a
sparse matrix. It represents the following matrix:
|
 |
Bond table (btab):
[0, 1, 0, 0, 0]
[1, 0, 2, 3, 0]
[0, 2, 0, 0, 0]
[0, 3, 0, 0, 4]
[0, 0, 0, 4, 0]
|
Please note that instead of getting the full bond table we are planning
to introduce a new method that gives back the bond index between atoms
i and j.
Chemically relevant information in the graph:
- number of connected fragments,
- rings: SSSR (Smallest Set of Smallest Rings)
Connections between the atoms specify the topology, but the relative
spatial arrangement of atoms – the configuration – also needs to be
defined, as molecules with same connectivity but different spatial
arrangement – stereoisomers
(see stereochemistry for details)
– should be expressed.
Spatial dimension of the building atoms defines the dimension of the
molecule.
- 0D – all atoms are in [0, 0, 0].
- 2D – z coordinate is 0, [x, y, 0].
- 3D – all coordinates are defined [x, y, z]
The dimensionality can be retrieved by the getDim()
method of MoleculeGraph class. It can be set by the
setDim(int d) method, but setDim(int d)
does not generate coordinates for the molecule. To generate coordinates,
which is obviously needed (bear in mind that it can be a slow process),
the clean(MoleculeGraph mg, int dim, java.lang.String opts)
method of chemaxon.calculation.clean.Cleaner
class can be used.
Once the molecule is built the atom and bond indexes are fixed,
they don't change with property changes (like changes in atom or
bond type), but if the structure is modified with removal/addition
of atoms or bonds, the indexes are changed.
Please note, that atoms and bonds are indexed from 0,
but MarvinView and MarvinSketch show the indexes starting from 1.
