Stereo Analysis - calculating stereo descriptors
Stereochemistry involves the study of the relative spatial arrangement of atoms and their transformations.
Stereochemistry defines different isomerisms that describe the molecule's behaviour in the space.
These covers the entire space of organic molecules. We do not give further details on stereochemistry here, instead
we refer to Marvin's stereochemistry summary:
http://www.chemaxon.com/marvin/help/sci/stereo-doc.html
It can be important to determine a molecule's stereochemical behaviour. This can be achieved by calculating stereochemical
descriptors for the molecule.
ChemAxon's stereo analysis module is able to calculate stereochemical descriptors for a molecule, giving an analysis in terms of the stereochemical properties.
The stereo analysis functionality is built in different ChemAxon products.
Stereo analysis with cxcalc
The solubility predictor is integrated into the cxcalc command line tool. The command syntax is
cxcalc [general options] [input files/strings] stereoanalysis [stereoanalysis options] [input files/strings]
where the stereoanalysis options are the following:
stereoanalysis options:
-h --help this help message
-T --type stereo descriptor type [tetrahedral | cistrans |
axial | atrop] (default: not set)
Some examples on how to use the stereo analysis module via cxcalc:
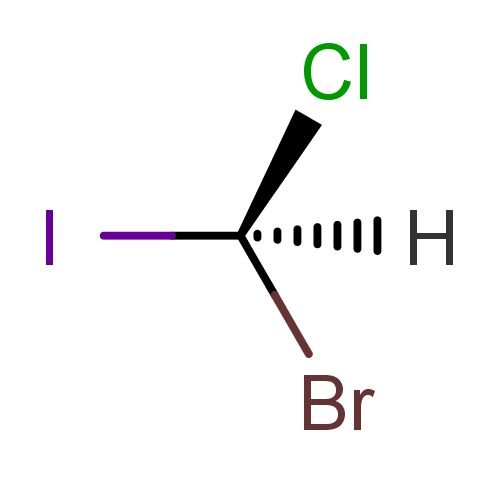
- Calculating tetrahedral stereo descriptors for the following molecule:
cxcalc stereoanalysis "[H][C@](Cl)(Br)I"
The output shows the calculated tetrahedral stereo descriptors:
TETRAHEDRAL [1] - [0, 2, 3, 4] : EVEN
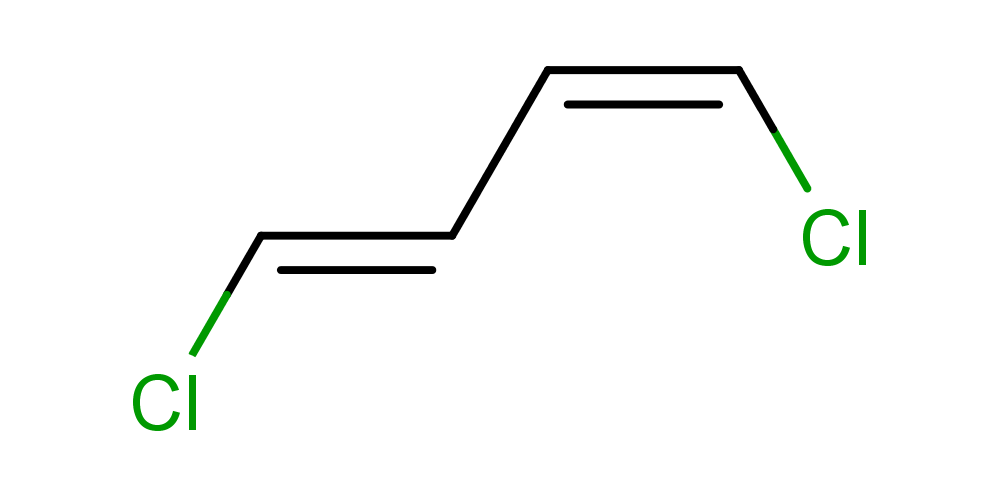
- Calculating cis-trans stereo descriptors:
cxcalc stereoanalysis "Cl/C=C\C=C\Cl"
The output shows the calculated cis-trans stereo descriptors:
CISTRANS [1, 2] - [0, 3] : CIS
CISTRANS [3, 4] - [2, 5] : TRANS
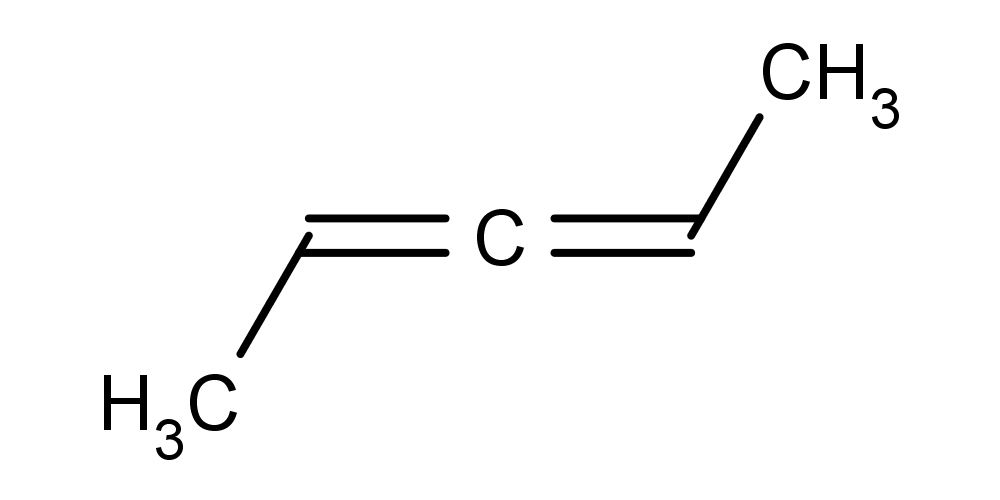
- Calculating axial stereo descriptors for a molecule:
cxcalc stereoanalysis CC=C=CC
The output shows the calculated axial stereo descriptors:
AXIAL [1, 3] - [0, 4] : WIGGLY
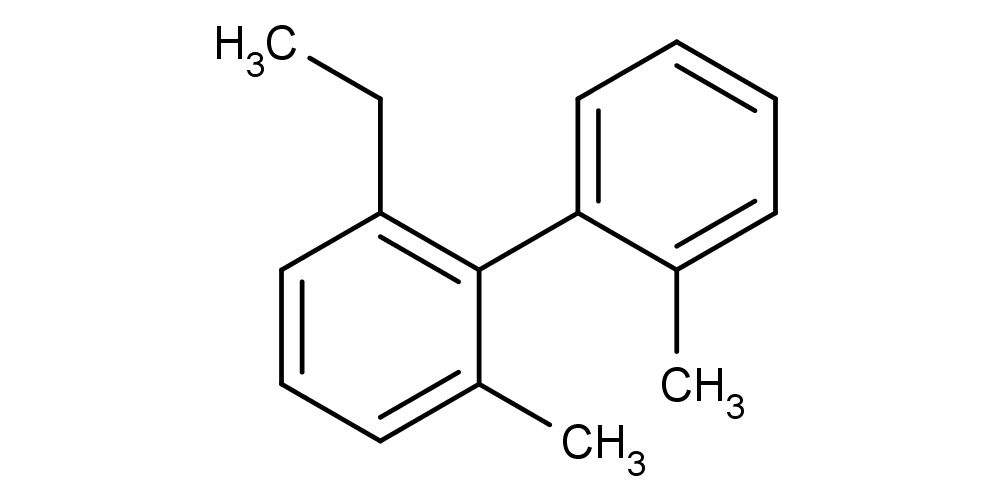
- Calculating atrop stereo descriptor for a molecule:
cxcalc stereoanalysis "CCC1=C(C(C)=CC=C1)C1=CC=CC=C1C"
The output shows the calculated atrop stereo descriptors:
ATROP [3, 9] - [4, 2, 14, 10] : UNKNOWN
- Calculating all stereo descriptors for a molecule with different stereo features:
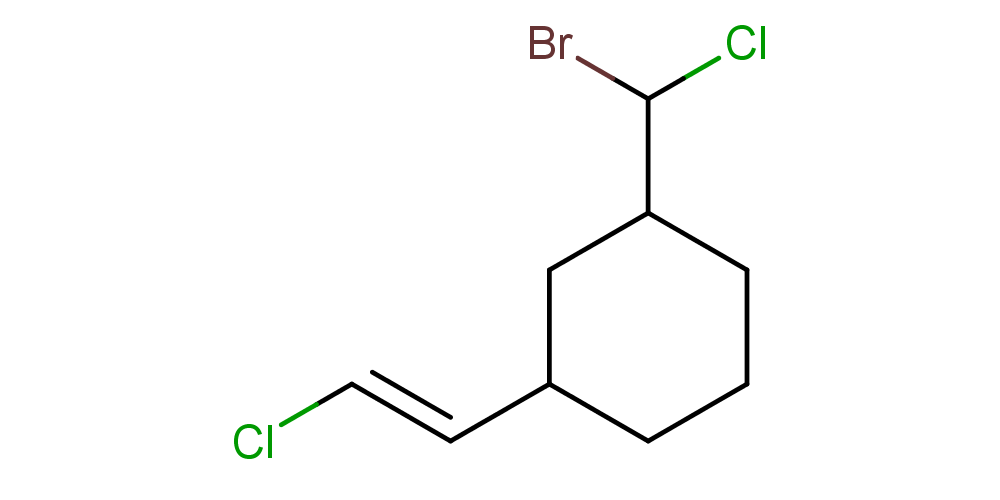
cxcalc stereoanalysis "ClC(Br)C1CCCC(C1)/C=C/Cl"
The output shows the calculated stereo descriptors:
CISTRANS [9, 10] - [7, 11] : TRANS
TETRAHEDRAL [3] - [1, 4, 8] : UNKNOWN
TETRAHEDRAL [1] - [0, 2, 3] : UNKNOWN
TETRAHEDRAL [7] - [6, 8, 9] : UNKNOWN
- Listing just the cis-trans stereo descriptors for the previous molecule:
cxcalc stereoanalysis -T cistrans "ClC(Br)C1CCCC(C1)/C=C/Cl"
The output is:
CISTRANS [9, 10] - [7, 11] : TRANS
Stereo analysis with ChemAxon's Chemical Terms
The stereo analysis module is also integrated into ChemAxon's Chemical Terms language.
The stereo descriptors can be calculated by two different functions in Chemical Terms:
- stereoanalysis(): calculates and returns the stereo descriptors for the input molecule.
- stereoanalysis(T): calculates and returns the stereo descriptor of type T.
The following examples show how the functions above can be used with the Chemical Terms evaluator
command line tool: