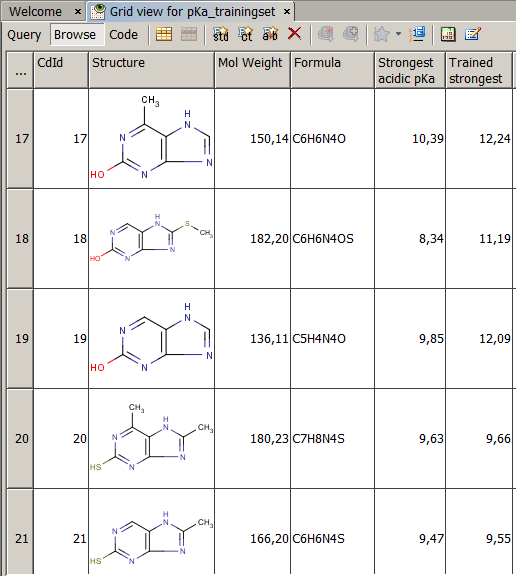
A sample of a typical training set is shown in the picture (pKa_trainingset.sdf). ID1 is the index of the atom with the experimental pKa1 value.
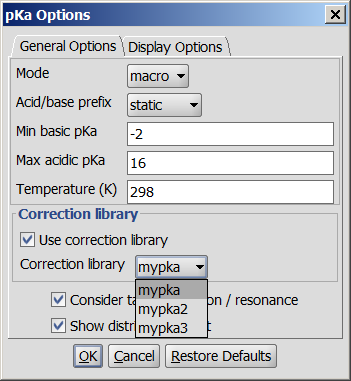
If you think your experimental data improved the accuracy of the default pKa calculator, you can take advantage of the supervised pKa learning method that is built into the pKa calculator. Special structural parts can have an effect on the pKa values calculated by the built-in method, so your correction library based on your experimental data helps the pKa calculator increase the prediction accuracy.
Inaccurately predicted ionization centers need to be identified and experimental data for them have to be collected. Since the learning algorithm is based on linear regression analysis, therefore you need to collect as many experimental pKa data as possible otherwise there won't be correlation. There is no strict rule on the number of the experimental datapoints, if your purpose is to create a local model, only for a certain type of chemical of the ionization centers, then it may be enough to collect a few representative structures. A robust model, however, requires as many diverse structures and pKa values as possible.
The experimental data should be collected in an sdf file, then the training algorithm has to be run which creates a correction library. This will be stored on your local computer, in your user folder. Finally, this correction library via MarvinSketch, cxcalc, Chemical Terms can be applied.
Input file preparation
A sample of a typical training set is shown in the picture (pKa_trainingset.sdf). ID1 is the index
of the atom with the experimental pKa1 value.
cxtrain pka -i [library name] [training file]Example:
cxtrain pka -i mypka mydata.sdf
|
|
| I. pKa calculation with training data | II. pKa calculation without training data |
|---|
--correctionlibrary or its short form: -L.
cxcalc pKa --correctionlibrary [library name] [input file/string]
Example
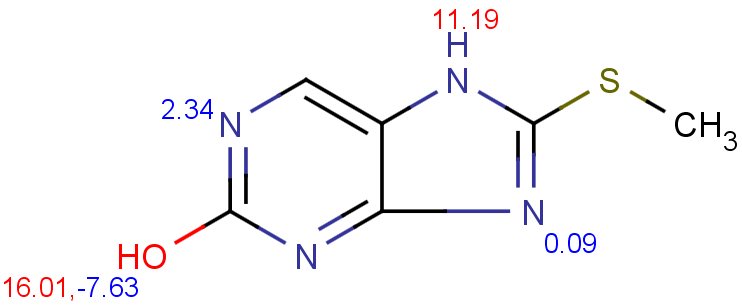
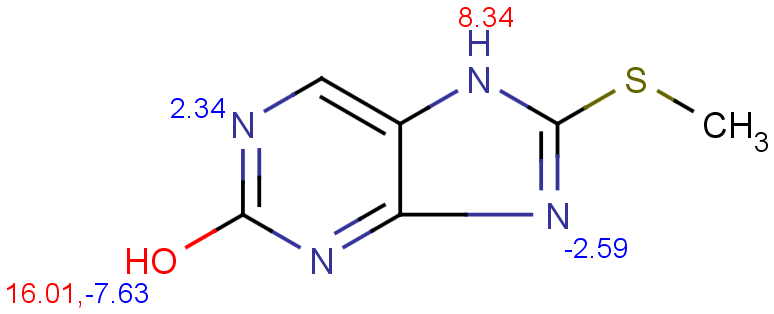
$ cxcalc pKa --correctionlibrary mypka "CSC1=NC2=C(N1)C=NC(O)=N2"
Result
id apKa1 apKa2 bpKa1 bpKa2 atoms
1 11.19 16.01 2.34 -2.59 7,11,9,4
$ cxcalc pKa "CSC1=NC2=C(N1)C=NC(O)=N2"Result
id apKa1 apKa2 bpKa1 bpKa2 atoms
1 8.34 16.01 2.34 -2.59 7,11,9,4
For more options see this page.
evaluate -e "pKa('correctionlibrary:[library name]')" "[input file/string]"evaluate -e "pKa('correctionlibrary:mypka')" "CSC1=NC2=C(N1)C=NC(O)=N2";;;-2,59;;;11,19;;2,34;;16,01;For more evaluator functions on pKa training see this page.
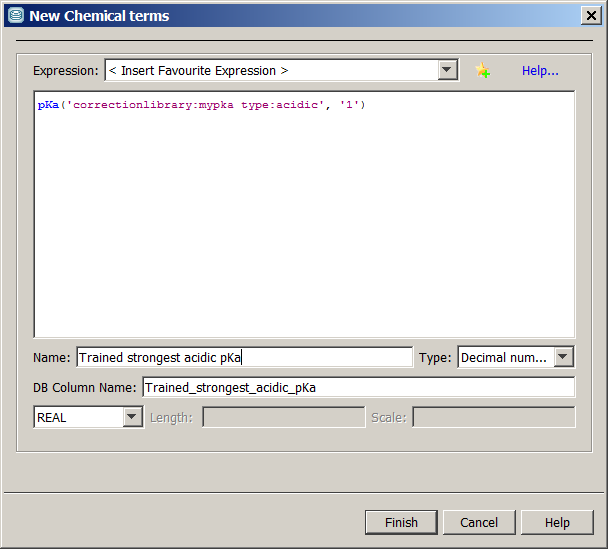
pKa ('correctionlibrary:mypKa type:acidic','1') defines that the plugin use the correction library named mypKa, and it will calculate the strongest acidic pKa of the molecule(s).