Isomers
Tautomers are organic compounds that are interconvertible by tautomerization.
Tautomerization reaction results in the formal migration of a hydrogen atom or proton,
accompanied by a switch of a single bond and adjacent double bond. Commonly, the
catalysts of these reactions are acids or bases. In solution a chemical equilibrium
of the tautomers will be reached. Some types of tautomers: ketone-enol, amid-imidic
acid, lactam-lactim, enamine-imine. Learn more about tautomerization and tautomers.
Tautomers of a compound can be determined with the help of Tautomer Generator Plugin.
Note: Tautomer Generator Plugin does not consider the three dimensional structure of molecules
during tautomer generation, and symmetric structures are filtered out from the generated tautomer set.
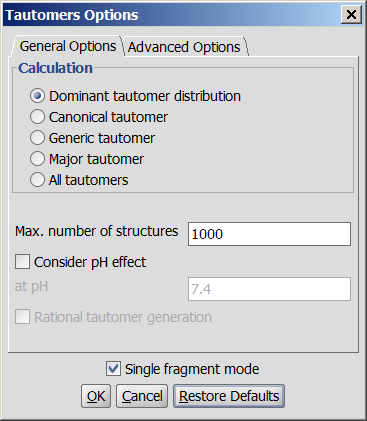
Following options can be adjusted in the Tools > Isomers > Tautomers,
Tautomers Options panel:
General options
- Calculation:
- Dominant tautomer distribution: displays the percentage of different tautomers present at the given pH.
- Canonical tautomer: calculates only the canonical tautomer of the structure. Rational tautomer generation mode can be activated.
- Generic tautomer: used for the identification of tautomers in JChem databases. It is calculated according to these rules:
- Tautomeric regions are identified.
- All bond types in the tautomeric regions will be changed to ANY.
- Each region will be assigned a data S-group with Sum(bonding electrons).
- Explicit hydrogens are removed.
- Isotope hydrogen:
- outside of tautomer regions is kept as is
- inside tautomer regions:
- Non-mobilizable isotope hydrogen (attached to an atom which is neither donor nor acceptor, so does not lose or gain H during tautomerization): the isotope is kept as is.
- Mobilizable isotope hydrogen (attached to a donor or acceptor atom in the tautomer region):
the mobilizable isotopic hydrogens are removed, and the number of each type is included in the data sgroup description. For example: "36 e 2 D 3 T" (meaning 36 bonding electrons, 2 tautomerizable Deuterium and 3 Tritium atoms in the region).
- Only the protection or deprotection of tetrahedral stereo centers is taken into consideration.
- Major tautomer: gives the first species from the dominant tautomer distribution.
- All tautomers: calculates all possible tautomers. If any deuterium or tritium is involved in the tautomerization, it moves during enumeration. Rational tautomer generation mode can be activated.
Max. number of structures: maximize the number of structures to display. This number
is the sum of unique tautomeric count and degenerated tautomeric count; however, only unique tautormers are displayed their degenerated tautomeric pairs are not.
Consider pH effect: takes into account the protonation states at given pH. Applicable for
Major tautomer and Dominant tautomer distribution calculations.
Rational tautomer generation: the tautomerization products are generated according to empirical rules. Rational tautomer generation narrows down the possible tautomerization paths and leads to chemically more feasible products.
Single fragment mode: if checked (default), the results are displayed in separate windows; if unchecked, the calculation handles unlinked molecules together and results are in the same window.
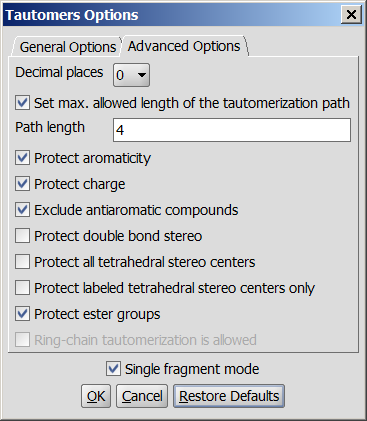
Advanced options
Note: the number of generated tautomers strongly depends on the options chosen.
- Decimal places: setting the number of decimal places with which
the tautomer distribution values are given.
- Set max. allowed length of the tautomerization path; Path length:
sets the number of bonds which are considered by displacing a double bond.
- Protect aromaticity: if checked (default), the aromaticity will be maintained.
- Protect charge: if checked (default), defined charged atoms maintain their charge during calculation.
- Exclude antiaromatic compounds: if checked (default), any tautomer structure having an
antiaromatic ring system will be discarded.
- Protect double bond stereo: if checked, all double bonds with stereo information
remain intact. If unchecked (default), tautomer regions will lose the double
bond stereo information, any other
stereo information in the molecule is kept intact.
- Protect all tetrahedral stereo centers: if checked, stereocenters are not
included in the tautomerization. If unchecked (default), tautomer regions will lose the tetrahedral
stereo information, any other
stereo information in the molecule is kept intact.
- Protect labeled tetrahedral stereo centers only: if checked, stereocenters
labeled with chiral flag or MDL Enhanced Stereo Representation flags will
not be included in tautomerization, other stereocenters will.
- Single fragment mode: if checked (default), the results are displayed in
separate windows, if unchecked, the calculation handles unlinked molecules together and
results are in the same window.
- Protect ester groups: if checked, ester is not taking part in tautomerization.
- Ring-chain tautomerization is allowed: this option can be activated when "All tautomers" function is selected. If it is checked, tautomer generation will take into account the possibility of ring closure.
For example, the following structures are the
calculated tautomers of 4-amino-6-ethoxypyrimidin-2-ol:
| Dominant tautomer distribution | 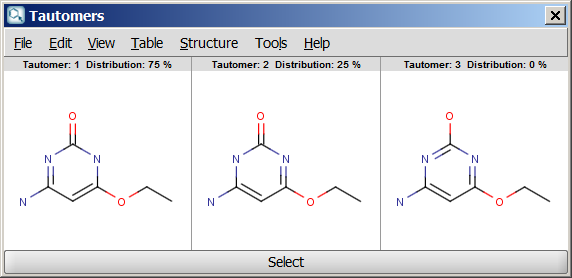
|
| Canonical tautomer | 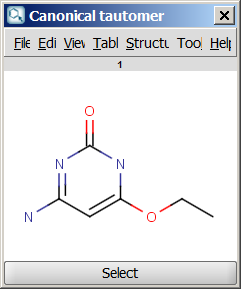
|
| Generic tautomer and an isotope labeled example | 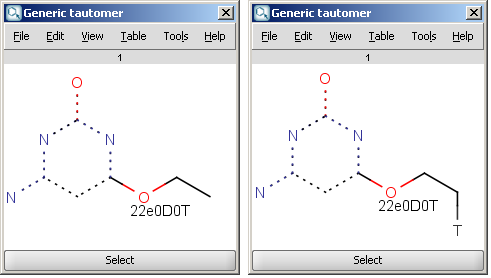
|
| Major tautomer | 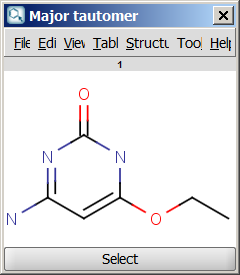
|
| All tautomers | 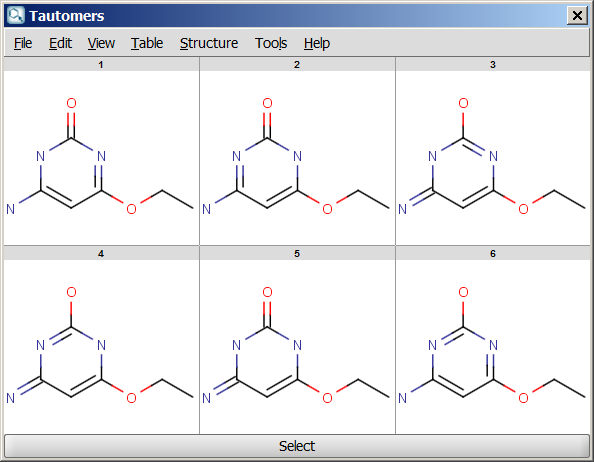
|
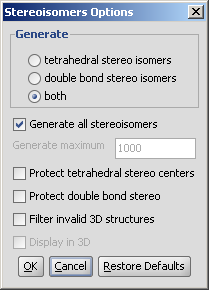
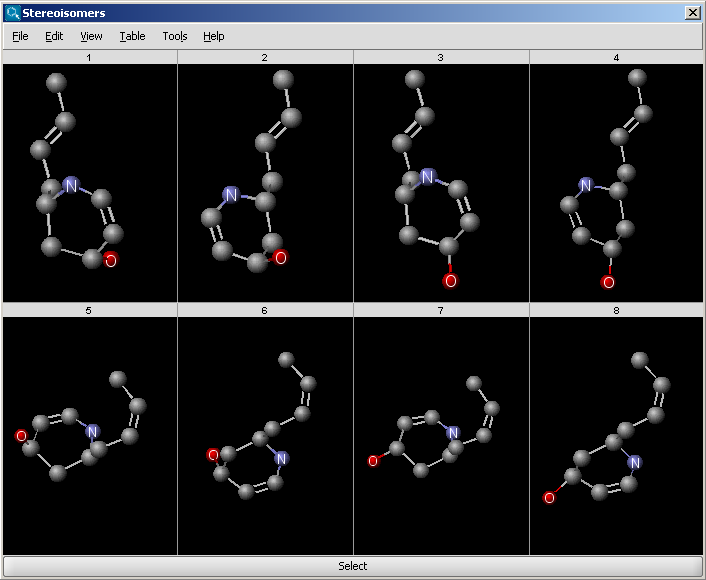
The Stereoisomers Generator Plugin produces all possible stereoisomers of a
given compound. The plugin handles both tetrahedral and double bond
stereo centers.
- Generate
- Tetrahedral stereo isomers: only the R/S isomers are generated.
- double bond stereo isomers: only E/Z isomers are generated.
- both: both R/S and E/Z isomers are generated.
- Generate all stereoisomers: all isomers are generated
- Generate maximum: only the given number of structures are generated.
- Protect tetrahedral stereo centers: if checked, preset stereocenters are not included in the stereoisomer generation.
- Protect double bond stereo: if checked, all double bonds with preset stereo information remain intact.
- Filter invalid 3D structures: sterically restricted isomers are discarded.
- Display in 3D: results are displayed in a 3D viewer.
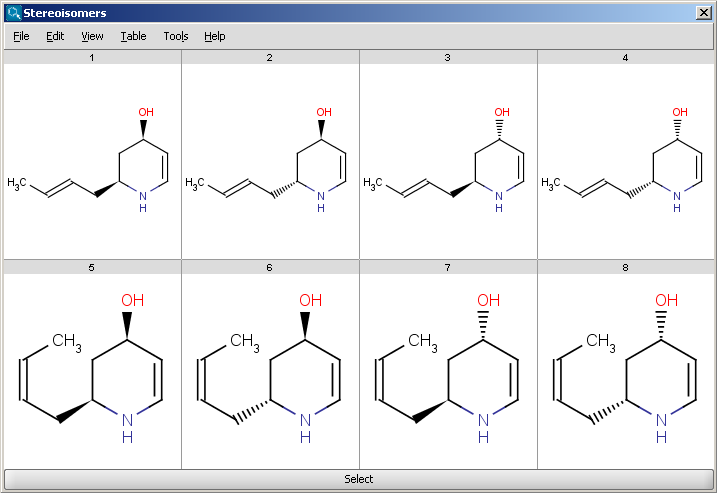
Results are displayed in a 2D viewer by default:
To replace your drawn molecule in the sketcher with any of the isomers shown, click on the structure then press "Select" at the bottom of the cells (the result window will be closed).
If "Filter invalid 3D structures" option is switched on in the
Stereoisomers Options panel, the stereoisomers can also be displayed in 3D.
References
- Smith, M. B.; March, J. Advanced Organic Chemistry, 5th ed., Wiley Interscience, New York, 2001; pp 1218-1223. ISBN 0471585890